Histone Modification Types and Effects
This Brush Up is sponsored by Cayman Chemical. Learn more about chemical inhibitors of histone modification.
Histone modification affects how tightly DNA wraps around histone proteins, yielding epigenetic changes that may activate or inhibit gene expression by influencing a gene’s availability to the cellular transcription machinery. This article explores various types of histone modifications, their biological effects, and how scientists develop compounds targeting these modifications to treat various diseases.
Histone modification can alter the nucleosome, a basic structural unit of chromatin that influences gene expression and cellular processes.
iStock, 7activestudio
What Is Histone Modification?
Histone modification refers to posttranslational alterations on histone proteins, H1, H2A, H2B, H3, and H4.1 The addition or removal of various chemical groups at specific amino acid residues influences how histone proteins and DNA interact within chromatin subunits called nucleosomes, where the DNA double helix wraps around the core histone octamer.
Modification of histones may result in chromatin remodeling or incomplete DNA unwinding, affecting chromatin structure and gene expression. Similar to epigenetic regulation via DNA methylation, posttranslational histone modifications do not alter the DNA sequence but can change its availability for transcription.2
Histone Modification Types
Most histone modifications occur within the amino terminus or N-terminal tail regions of histone proteins, which are unstructured and participate in DNA interactions that modulate several nucleosome dynamics such as positioning and stability.3 Different catalytic enzymes, such as histone acetyltransferases (HATs), histone methyltransferases (HMTs), histone deacetylases (HDACs), and histone demethylases (HDMs), can chemically modify histones via posttranslational processes.4
Histone acetylation and deacetylation
Histone acetylation is an epigenetic modification that plays a crucial role in transcription regulation and DNA repair.5 During the histone acetylation process, HATs utilize acetyl co-enzyme A (acetyl-CoA) to catalyze a chemical reaction that adds an acetyl group to lysine residues. This process neutralizes the lysine’s positive charge, weakening its DNA interactions.
In contrast, HDACs remove acetyl groups from histone lysine residues, tightening DNA interactions and forming a more condensed chromatin structure. The deacetylation process also blocks transcription factor binding, inhibiting gene expression. Scientists have observed that both histone acetylation and deacetylation regulate transcription in numerous cell types, including stem cells and cancer cells.
Histone methylation and demethylation
Histone methylation is a dynamic process that promotes transcriptional repression and, in some cases, transcriptional activation.6 Specialized HMTs add methyl groups to different amino acid residues. For example, protein arginine methyltransferases (PRMTs) typically add methyl groups to arginine and histone lysine methyltransferases (HKMTs) modify lysine.
Conversely, HDMs remove methyl groups from histone proteins via demethylation. Experimental studies have shown that both histone methylation and demethylation processes play a crucial role in maintaining genome integrity and proper tissue and organ development.7,8
Histone phosphorylation and dephosphorylation
Histone phosphorylation is a posttranslational alteration that influences chromatin remodeling, DNA repair, and transcription. Histone kinases typically catalyze the transfer of a phosphate group from adenosine triphosphate (ATP) to the hydroxyl group of a specific histone serine, threonine, or tyrosine residue. Scientists have identified roles for histone phosphorylation in chromatin remodeling, in addition to a well-known DNA damage response role via phosphorylated histone variant H2AX.9
Both phosphorylation and dephosphorylation processes influence DNA packaging by altering the charge of histones. In turn, these changes affect diverse biological events, including transcription, apoptotic responses, and chromosome segregation.10 In general, histone phosphorylation results in a more open and accessible chromatin structure, while dephosphorylation typically leads to chromatin condensation.
Histone ubiquitination and deubiquitination
Histone ubiquitination is another reversible posttranslational modification that alters transcriptional states and DNA integrity; however, unlike modifications that add chemical groups to histones, ubiquitination adds a small protein called ubiquitin.11 Histone ubiquitination involves a series of sequential catalytic reactions via ubiquitin-activating and conjugating enzymes, as well as ligases, that form a bond between ubiquitin’s carboxy-terminal (C-terminal) glycine residue and a lysine residue on a histone tail or another ubiquitin that is already attached to the histone.
Peptidases are responsible for deubiquitinating histones and other proteins. The balance between ubiquitination and deubiquitination is crucial for numerous cellular processes such as DNA repair, cell cycle progression, and signal transduction.
Biological Significance of Histone Modification and Epigenetics
Histone modifications play a vital role in many biological processes, including cellular replication pathways such as meiosis, an integral part of gametogenesis.12 For instance, during meiosis, the chromatin structure undergoes dynamic changes, and modification of histones is among several mechanisms that ensure proper chromatin dissolution and reorganization. As such, histone modification is crucial for normal development and distinct cell lineage differentiation in adult organisms. Additionally, histone modification influences nearly all DNA-based processes, including replication, transcription, recombination, and DNA damage repair.13
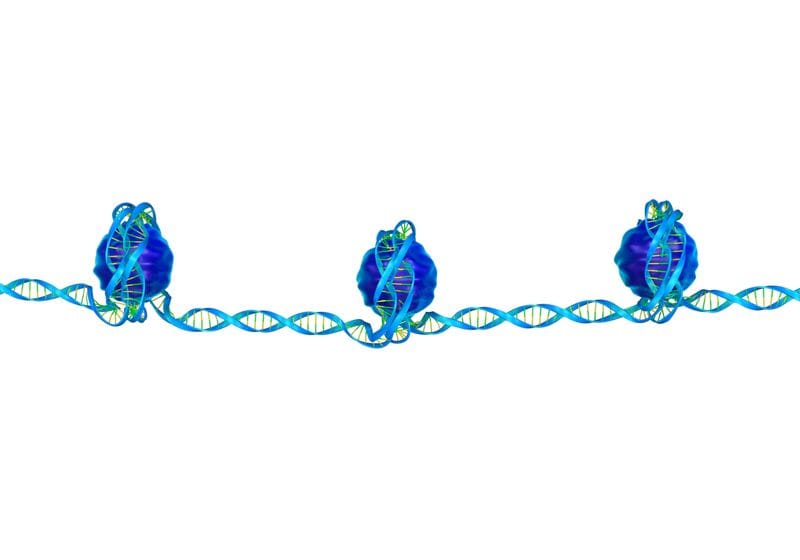
Histone modification on the tails of different histone proteins can play a variety of biological roles, regulating transcription, chromatin remodeling, the DNA damage response, and beyond.
Modified from © iStock, KKT Madhusanka, ttsz
Table: Biological significance of histone modification
Histone Modification Type | Enzymes | Example Functions |
Acetylation14 | Acetyltranferases (e.g., GNAT, MYST, and CBP/p300) | Promotes open chromatin conformation and gene activation |
Deacetylation14 | Deacetylases (e.g., class I HDAC, class II HDAC, and class IV HDAC) | Promotes chromatin condensation and gene repression |
Methylation14 | Lysine methyl-transferases (e.g., NSD1) | Promotes gene silencing and gene activation |
Demethylation14 | Lysine demethylases (e.g., LSD1) | Regulates gene expression during development |
Phosphorylation15 | Kinases (e.g., Aurora B kinase) | Promotes chromosome condensation during mitosis |
Dephosphorylation16 | Phosphatases (e.g., EYA) | Regulates repair or apoptotic factor recruitment, influencing whether a cell repairs DNA damage or undergoes cell death |
Ubiquitination17 | E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases | Promotes gene activation |
Similar to other epigenetic mechanisms, histone modification plays a role in several disease processes.18 Researchers have linked abnormal changes in histone modifier enzyme activity to cancer initiation, progression, and metastasis; neurodegenerative diseases; and developmental disorders. For example, histone-encoding gene mutations at codons for commonly methylated lysine residues are associated with increased cancer risk.19 In children, lysine to methionine substitutions at residues 36 and 27 within H3 (H3K36M and H3K27M) cause chondroblastoma and diffuse intrinsic pontine glioma (DIPG), respectively.
Chemical Inhibition of Histone Modifications as a Therapeutic Strategy
Technological advances have allowed researchers to study histone modification for translational applications. For example, chromatin immunoprecipitation sequencing technology enables precise mapping of histone methylation patterns at specific genomic regions.20 This precision has helped scientists design and develop targeted chemical inhibitors that can alter histone modification.
Before therapeutic chemical inhibitors progress to the clinic, scientists comprehensively assess their safety profiles and off-target consequences. Because histone modification is a balancing act of multiple reversible reactions rather than a therapeutic on-off switch, researchers investigate how inhibiting a modification in one context might tip the balance of another. For example, cellular studies have suggested that, in some instances, HDAC inhibitors participate in the pathogenesis of neurodegenerative diseases, while in other cases they show therapeutic potential. This could be due to the varying epigenetic status of the targets, cell types, or tissue specificity.21
Examples of chemical inhibitors
Researchers have developed numerous inhibitors that target lysine acetyltransferase (KAT) activity, primarily focusing on GNAT/PCAF, p300/CBP, and MYST classes. These inhibitors typically target the enzymatic activity and acetyl-CoA binding sites. CCS1477 is another type of small molecule inhibitor that targets the bromodomains of p300 and CBP, which are protein domains that recognize acetylated lysine residues. The CBP and p300 bromodomains play crucial roles in cancer cell growth and survival, and initial research findings indicate their potential as therapeutic targets in hematological malignancies and prostate cancer.22
Scientists have also determined that abnormal expression patterns of HMTs such as SMYD3 and EZH2 promote various types of cancer, including breast and oral cancer.23 The US Food and Drug Administration (FDA) has approved several drugs targeting histone modifications, including tazemetostat, an oral EZH2 inhibitor, which is indicated to treat follicular lymphoma and epithelioid sarcoma.24
Besides cancer, researchers have also revealed the neuroprotective role of HDAC inhibitors in treating various central nervous system (CNS) disorders.21 Experimental findings indicate the efficacy of HDAC inhibitors (e.g., hydroxamates (vorinostat), belinostat (PXD101), panobinostat (LBH589) and trichostatin A) for the treatment of neurological disorders, including Huntington’s disease, amyotrophic lateral sclerosis, Alzheimer’s disease, spinal muscular atrophy, Rubinstein-Taybi syndrome, and Fragile X syndrome.
Final Thoughts on Histone Modification
Considering the significant benefits of histone modification therapies, scientists seek to discover novel small-molecule compounds that can modify histone proteins for therapeutic purposes. Future research also aims to improve treatment effectiveness and minimize side effects by identifying new targets and enhancing drug specificity.
FAQ
What is histone modification?
- Histone modification refers to posttranslational alterations on histone proteins, typically through the addition or removal of chemical groups or small protein modifiers at specific amino acid residues in the histone tail regions.
What is the purpose of histone modification?
- Histone modification influences how histone proteins and DNA interact within chromatin subunits called nucleosomes, where the DNA double helix wraps around the core histone octamer. Modification of histones can regulate nearly all DNA-based processes, including chromatin remodeling, gene expression, and the DNA damage response.
What is the difference between DNA methylation and histone methylation?
- DNA methylation and histone methylation are both epigenetic modifications that affect gene expression without altering the DNA sequence. DNA methylation involves the addition of methyl groups to DNA bases, while during histone methylation, specialized enzymes add methyl groups to amino acid residues such as lysine and arginine.
- Zhang K, et al. Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE. 2007;2(11):e1210.
- Alaskhar Alhamwe B, et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin Immunol. 2018;14:39.
- Munshi A, et al. Histone modifications dictate specific biological readouts. J Genet Genomics. 2009;36(2):75-88.
- Marmorstein R, Trievel RC. Histone modifying enzymes: Structures, mechanisms, and specificities. Biochim Biophys Acta. 2009;1789(1):58-68.
- Dutta A, et al. Diverse activities of histone acylations connect metabolism to chromatin function.Mol Cell. 2016;63(4):547-552.
- Song Y, et al. Targeting histone methylation for cancer therapy: Enzymes, inhibitors, biological activity and perspectives. J Hematol Oncol. 2016;9(1):49.
- Mushtaq A, et al. Role of histone methylation in maintenance of genome integrity.Genes (Basel). 2021;12(7):1000.
- Jorgensen BG, Ro S. Role of DNA methylation in the development and differentiation of intestinal epithelial cells and smooth muscle cells. J Neurogastroenterol Motil. 2019;25(3):377-386.
- Rossetto D, et al. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7(10):1098-108.
- Aboulache BL, et al. Phosphorylation regulates the chromatin remodeler SMARCAD1 in nucleosome binding, ATP hydrolysis, and histone exchange.J Biol Chem. 2024;300(12):107893.
- Wang J, et al. Ubiquitin regulation: The histone modifying enzyme’s story. Cells. 2018;7(9):118.
- Nie H, et al. Roles of histone post-translational modifications in meiosis. Biol Reprod. 2024;110(4):648-659.
- Yao W, et al. Crossing epigenetic frontiers: The intersection of novel histone modifications and diseases. Signal Transduct Target Ther. 2024;9(1):232.
- Handy DE, et al. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation. 2011;123(19):2145-56.
- Goto H, et al. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002;7(1):11-7.
- Cook PJ, et al. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458(7238):591-6.
- Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26.
- Yang Y, et al. The roles of histone modifications in tumorigenesis and associated inhibitors in cancer therapy. J Natl Cancer Cent. 2022;2(4):277-290.
- Wan YCE, et al. Histone H3 mutations in cancer. Curr Pharmacol Rep. 2018;4(4):292-300
- Satam H, et al. Next-generation sequencing technology: Current trends and advancements. Biology (Basel). 2023;12(7):997.
- Shukla S, Tekwani BL. Histone deacetylases inhibitors in neurodegenerative diseases, neuroprotection and neuronal differentiation. Front Pharmacol. 2020;11:537.
- Gou P, Zhang W. Protein lysine acetyltransferase CBP/p300: A promising target for small molecules in cancer treatment. Biomed Pharmacother. 2024;171:116130.
- Liu R, et al. Post-translational modifications of histones: Mechanisms, biological functions, and therapeutic targets. MedComm (2020). 2023;4(3):e292.
- Straining R, Eighmy W. Tazemetostat: EZH2 inhibitor. J Adv Pract Oncol. 2022;13(2):158-163.